
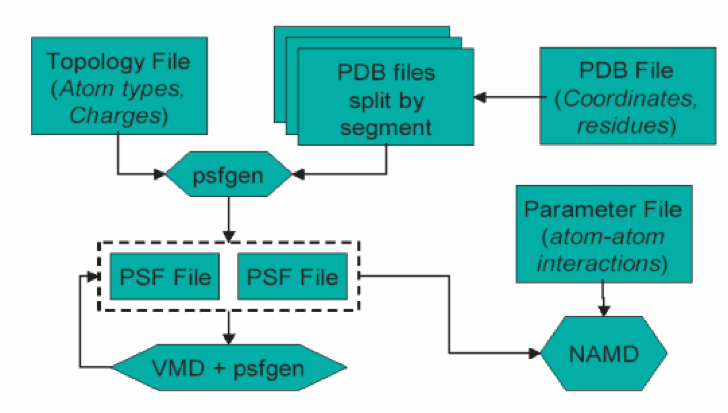
Dabble writes an input file for psfgen, and handles all its quirks (such as not allowing segments to have more than 10,000 atoms because its PDB parser is the worst). In TkConsole, enter the following script: Problem with solvate and PSF files in CHARMM format. The VMD plugin psfgen is used to generate parameterized files with CHARMM parameters. > psfgen) duplicate topology file > psfgen) Unable to add (duplicate). Next, in vmd, select Extension and Tk console. How do I create a PSF file from PDB First, load your pdb file (say water.
Vmd psfgen portable#
Do the same for the second (y coordinate) and third (z coordinate) numbers of each set. From: Isuru Herath () Date: Sun 08:56:47 CST Next message: Isuru Herath: 'Re: Readpsf Error'. The psfgen plugin is a structure building tool consisting of a library of portable structure and file manipulation routines that implement Tcl interface, callable from within VMD. Without taking signs (only absolute values), add the first number of the first set to the first number of the second set. Copy down the numbers that are outputted. Go to Extensions > TkConsole and enter the following script: Open VMD and load the ionized PSF, then the ionized PDB into the PSF. Psfgen is a structure building tool with a TCL interface commonly used as a VMD plugin to set up simulations using NAMD or other molecular dynamics software. psfgen is licensed by the University of Illinois Non-Exclusive, Non-Commercial use license. You will now need to change the Periodic Boundary Conditions. These can all be found in Drive under DDM > Topology Files.Į. Under "Simulation Parameters," make sure all of the parameter files are in the same directory as your micelle files. Change "set outputname" to micelle_min.ĭ. Under "Adjustable Parameters," change the structure to your ionized PSF and your coordinates to the ionized PDB. Change the title at the very top to reflect the appropriate micelle.Ĭ. Create a CONF file (you can use a previous researcher's minimization CONF file as a reference look for one ending with _min.conf) and save in the necessary directory.ī. You will now have a pdb and psf in your current directory with the prefix you chose in step e.Ī. Click Neutralize and set NaCl concentrate to (enter value) > Autoionize. Select the Input PSF and PDB to be the ones ending in _wb.psf and _wb.pdb if they are not already filled in.į. Change the Output prefix to an appropriate naming convention (for example, ionized_80mi for an 80-molecule micelle). Click Main > Extensions > Modeling > Add ionsĮ.

Delete the original PSF in the main console.ĭ. Solvate micelle.psf micelle.pdb -t 12 -o micelle_wbĬ. Click Extensions > TkConsole and enter the following script: Click browse again and load the PDB into the PSF. Go to File > New Molecule > Browse and select the PSF. This will create a PSF and a new pdb with correct coordinates of your micelle.Ī. Enter the following script (areas that are underlined and italicized indicate that these will need to be changed depending on your naming convention:Į. Click load and close the Molecule File Browser.ĭ. Click File > New Molecule > Browse and select the PDB. Open a terminal in this directory and type: vmdĬ. Navigate to the directory that contains your topology files and the PackMol PDB of your micelle.ī. Create a PSF and a new PDB of your micelle.Ī. To start, you must have the necessary topology files and the PDB of your micelle created in PackMol in the same directory.ġ. This guide also uses NAMD in conjunction with VMD. you can use a simple text editor to put.The following is a step-by-step guide for ionizing, solvating, and heating a micelle to prepare it for a run.
Vmd psfgen software#
The last TER designates termination of one chain, without it some software considers them as a single chain like your case. PDB format is composed of many columns but the first column contains the atom information like ATOM, HETATOM, or TER. However, when we have to put them back we have to follow PDB file format rules. To generate a topology file one has to generate each component separately, if PDB file has two chains then one has to separate them.
Vmd psfgen plus#
Plus little bitty understanding of the PDB file format and Structural Biology, and Python or Bash scripting skills. To simulate this interaction in silico, We need a proper 1) Forced field, 2) Topology files, and 3) Molecular Dynamic packages like NAMD, Gromacs, or Amber. These two proteins have strong binding energy therefore frequently used for studying protein-protein interaction. Barnase and barstar are the extracellular ribonuclease and its intracellular inhibitor produced by Bacillus amyloliquefaciens.


 0 kommentar(er)
0 kommentar(er)
